“We just found two more!” said tropical ecologist Dan Janzen. Although the telephone connection with him was shaky, his excitement was palpable. “The first butterfly two months ago, the other just two weeks ago.”
We had reached Janzen in January at the Area de Conservacion Guanacaste, a tropical forest preserve he helped create in northwestern Costa Rica, where Janzen does much of his widely lauded biodiversity research. (Among other honors, he has been awarded the John D. and Catherine T. MacArthur Foundation’s “genius” grant.) The “two more” were newly identified species of butterfly, both luminescent blue “skippers.”
Discovery of new species is reason enough for a biologist’s enthusiasm. But Janzen clearly was jazzed by something he sees as far more momentous — a technology called DNA barcoding that made these discoveries possible in the first place, and that promises to revolutionize the otherwise daunting process of identifying the millions of species on the planet, many yet unknown and unnamed.
The term “barcoding” is actually an analogy. Much the same way that a small universal product barcode allows a retailer’s scanners to distinguish a box of tissues from a can of green beans, DNA barcoding technology allows scientists to use data from a tiny snippet of a single gene to distinguish one species from the next. Although not perfect, proponents say it is highly accurate in distinguishing almost all species of animals, with a promising variation under development for plants. At a few dollars per species it is also remarkably cheap and, compared to traditional DNA analysis, lightning fast.
Eventually, it might even be possible to embed the technology into an inexpensive handheld device. When that happens, Janzen says, “it will do for biodiversity what the printing press did for literacy.” He envisions a gadget straight out of Star Trek, an electronic reader of the catalogue of life on the planet that would enable anyone — schoolteacher, farmer, curious child — to identify “what bit of biodiversity is biting them, appealing to them, worrying them” in an instant.
For now, DNA barcoding technology is limited to scientists with access to a few large labs with the right equipment. But those new butterfly species hint at the technology’s promise. The two new skipper species actually belong to a surprisingly large cluster of recently identified species that, for years, were hiding in plain sight. As recently as 2003, scientists thought that the butterflies cataloged under the name Astraptes fulgerator were all of a single species. The butterfly adults all seemed to look exactly the same, although, mysteriously, color patterns varied among their larval caterpillars. That year, Janzen, a biology professor at the University of Pennsylvania, gave tissue samples from dozens of butterflies his team had collected to geneticist Paul Hebert, of Ontario’s University of Guelph, who had developed the barcoding technology. His analysis quickly revealed not one, but what appear to be 10 distinct species.
The two new discoveries bring the total to 12, and there may even be a few more, Janzen says. Where science once saw a generalist species that occupied a wide variety of tropical habitats, DNA barcoding has uncovered an array of species that actually specialize and occupy differing habitats. Why all mimic the same color pattern remains a mystery, although it probably lends some sort of evolutionary advantage. Janzen thinks it’s likely a don’t-bother signal to would-be predators. Unusually fast fliers, skippers should lose any appeal to hungry birds that would learn as youngsters not to waste energy on hot pursuit of the hard-to-catch.
Beyond the arena of species discovery, the technology is rapidly finding its way into an array of practical applications, from helping health agencies to control insect-borne diseases, to helping airlines and the military avoid disastrous in-flight collisions with birds, to helping regulatory agencies monitor stream and lake quality.
Hebert says that although he’d been yearning for better ways to probe biodiversity during decades of field work in places as diverse as tropical New Guinea and the Canadian Arctic, the idea of DNA barcoding quite literally came to him as an out-of-the blue inspiration. One day in the late 1990s he was in a supermarket looking at retail barcodes when it hit.
“It occurred to me that if the retail industry can use a few numbers to represent a vast array of products, why can’t we look at DNA the same way?” he says. Herbert quickly set out to determine if he could find a genetic snippet common to all species that could yield enough information to tell them apart. He needed to find a bit of DNA small enough to be sequenced quickly. And it needed to be, like a bit of DNA Goldilocks might find, “just right.” The gene had to be one that mutates quickly enough to be distinct from that of a recent evolutionary ancestor. But it also had to mutate slowly enough that barcodes would not vary markedly within a species.
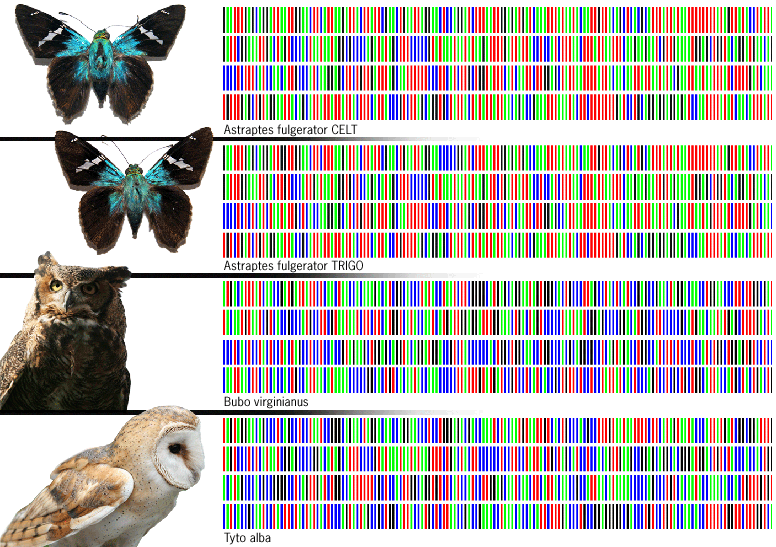
He found a just-right sequence on the first 648 bits of DNA of a gene called cytochrome c oxidase subunit 1 (COX1). Each long strand of DNA contains only four nucleotides: adenine, guanine, thymine, and cytosine. The complex patterns in which they are arranged determine whether your eyes are blue or brown, or whether an organism is a zebra or a zebra fish. Herbert found that by simply recording the precise order of DNA compounds, called nucleotides, on the COX1 gene, he would have a de facto barcode that would define a unique species. And it turns out that a color-coded chart of a unique A, G, T, and C sequence looks enough like a supermarket barcode to make the analogy surprisingly apt. The COX1 gene works well for animal species and at least some fungi and algae. Scientists developing barcoding technology for plants now believe that they may need to use DNA from two or three genes for accurate results.
“There was a lot of skepticism that we could deliver something that actually works,” Hebert says of the years early in this decade when he was largely frustrated in attempts to find support to pursue the technology. Some traditional taxonomists, whose core work has, since the time of Linnaeus, involved close visual comparison of species, were especially skeptical. In fact, in a few cases of hybrids and of species recently evolved from others, barcoding needs to be supported by more detailed genetic analysis or visual comparison. But it is useful in enough cases, including about 98 percent of animal species, that barcoding projects are now growing explosively.
In one major effort, the Natural History Museum in London and the Smithsonian Institution in Washington D.C. are collaborating on a project to barcode all of the world’s known mosquito species. That should lead to far better targeting of species that are vectors of malaria and other devastating diseases. The project, which began only in early 2007, has already solved a puzzle in South America. An apparent single species called Anopheles oswaldoi ranges across a wide area but, curiously, seemed to be a malaria vector only on some of its range. DNA barcoding revealed it to be four distinct species, only one of which appears to be a disease carrier. Future control programs can now focus on controlling the waterborne larvae of only the harmful species.
In May, 2007, the U.S. Food and Drug Administration used barcoding to issue a warning that a shipment of supposed monkfish from China actually appeared to be a species of toxic pufferfish. And the U.S. Air Force and the Federal Aviation Administration (FAA) have helped fund a Smithsonian sponsored barcoding of all North American bird species. On the pure discovery side, the study has given scientists new leads on deciding whether some subpopulations are their own species. But the military and FAA helped fund it for more immediately practical reasons. Scientists are now using blood and tissue samples collected from aircraft to try to better understand which birds pose collision risks and where. In terms of numbers of species barcoded so far, said Hebert in January, “We’re sitting at 35,000 species, and feeling pretty happy about that.” Today, almost all barcoding is being done at a handful of labs in North America, Herbert’s own barcoding “factory” at the University of Guelph, and a facility at the Smithsonian. But he says that if funding for the new multination International Barcode of Life (iBOL) coalition comes together, 500,000 of the world’s catalogue of species could be barcoded in another five years. That would be the first step in perfecting an iPOD-like species reader that would contain a miniature DNA sequencer along with a tiny memory chip crammed with millions of barcodes, or what Herbert calls “life on the planet in a box.”
Another technological leap is likely to help. The Guelph lab recently obtained funding from the government of Canada to purchase a pair of devices that will allow DNA analysis in huge volume at blinding speeds. Conventional equipment can now provide 96 DNA “reads” in about two hours, or about 400,000 barcode records in a year. The new device can provide those 400,000 records in a single two hour run. As the equipment comes on line, Hebert envisions a new era of “environmental barcoding” that sorts out diverse assemblages of species from big unsorted samples. Picture, he says, a kilogram of insects collected from a rainforest canopy. “We can mix them into a bug milkshake,” he says, “toss them into the hopper, and tell you in a couple of hours what the 1000 species you’ve collected are. Or we could hoover up a little bit of stream bottom, and quickly tell you what species are present.”